 NVIDIA released NVIDIA Clara Parabricks Pipelines version 3.5, adding a set of new features to the software suite that accelerates end-to-end genome sequencing analysis.
NVIDIA released NVIDIA Clara Parabricks Pipelines version 3.5, adding a set of new features to the software suite that accelerates end-to-end genome sequencing analysis.
NVIDIA recently released NVIDIA Clara Parabricks Pipelines version 3.5, adding a set of new features to the software suite that accelerates end-to-end genome sequencing analysis.
With the release of v3.5, Clara Parabricks Pipelines now provides acceleration to Google’s DeepVariant 1.0, in addition to a suite of existing DNA and RNA tools. The addition of DeepVariant to Parabricks Pipelines brings highly-accurate variant calling for both short- and long-read sequencing data to the community.
This new release also enables graphical reports of QC metrics from binary alignment map (BAM) files to variant call files (VCF). Researchers can use these graphical reports to better assess the quality of their sequencing data and the subsequent variant calling before moving the results for additional downstream analysis.
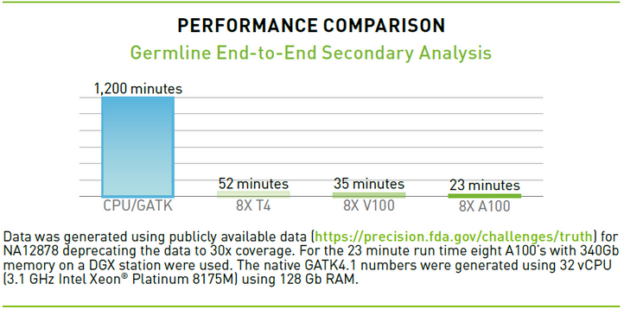
Parabricks Pipelines is packaged with enterprise support for A100 and other NVIDIA GPUs, offering one of the industry’s fastest compute frameworks for whole genome and whole exome applications. For a whole genome at 30x coverage, a server with 32 virtual CPUs takes about 1,200 minutes to generate a variant call file (VCF), while a server with eight A100 Tensor Core GPUs running Clara Parabricks takes less than 25 minutes to go from FASTQ to VCF.
Start a free one month trial of NVIDIA Clara Parabricks Pipelines today and learn how to get set up in just 10 minutes with this step-by-step instructional video.
