 The NVIDIA Swin UNETR model is the first attempt for large-scale transformer-based self-supervised learning in 3D medical imaging.
The NVIDIA Swin UNETR model is the first attempt for large-scale transformer-based self-supervised learning in 3D medical imaging. 
At the Computer Vision and Pattern Recognition Conference (CVPR), NVIDIA researchers are presenting over 35 papers. This includes work on Shifted WINdows UNEt TRansformers (Swin UNETR)—the first transformer-based pretraining framework tailored for self-supervised tasks in 3D medical image analysis. The research is the first step in creating pretrained, large-scale, and self-supervised 3D models for data annotation.
As a transformer-based approach for computer vision, Swin UNETR employs MONAI, an open-source PyTorch framework for deep learning in healthcare imaging, including radiology and pathology. Using this pretraining scheme, Swin UNETR has set new state-of-the-art benchmarks for various medical image segmentation tasks and consistently demonstrates its effectiveness even with a small amount of labeled data.
Swin UNETR model training
The Swin UNETR model was trained on an NVIDIA DGX-1 cluster using eight GPUs and the AdamW optimization algorithm. It was pretrained on 5,050 publicly available CT images from various body regions of healthy and unhealthy subjects selected to maintain a balanced dataset.
For self-supervised pretraining of the 3D Swin Transformer encoder, the researchers used a variety of pretext tasks. Randomly cropped tokens were augmented with different transforms such as rotation and cutout. These tokens were used for masked volume inpainting, rotation, and contrastive learning, for the encoder to learn a contextual representation of training data, without increasing the burden of data annotation.
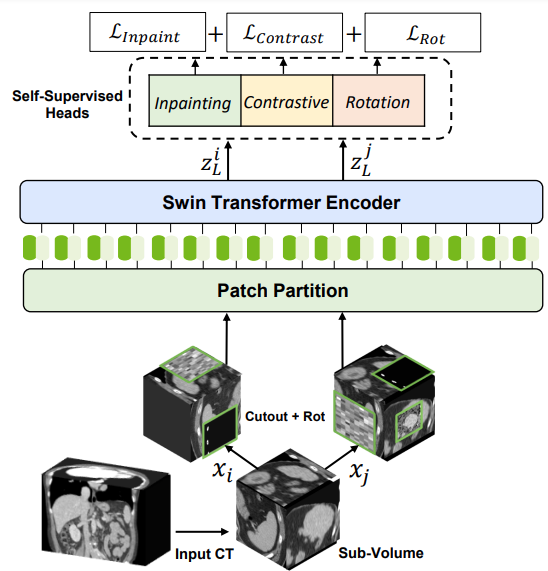
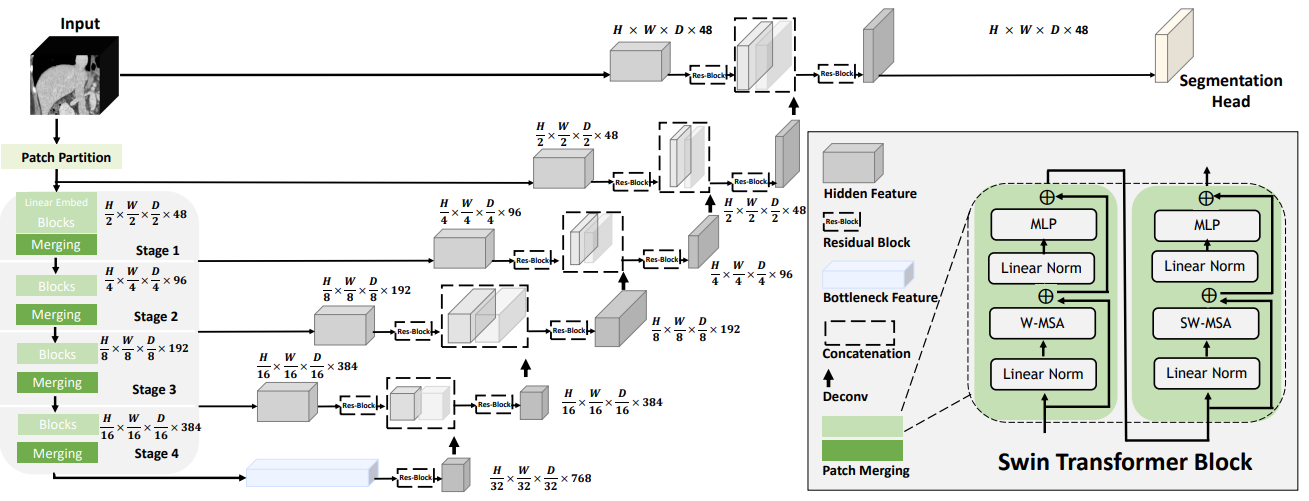
The technology behind Swin UNETR
Swin Transformers adopts a hierarchical Vision Transformer (ViT) for local computing of self-attention with nonoverlapping windows. This unlocks the opportunity to create a medical-specific ImageNet for large companies and removes the bottleneck of needing a large quantity of high-quality annotated datasets for creating medical AI models.
Compared to CNN architectures, the ViT demonstrates exceptional capability in self-supervised learning of global and local representations from unlabeled data (the larger the dataset, the stronger the pretrained backbone). The user can fine-tune the pretrained model in downstream tasks (for example, segmentation, classification, and detection) with a very small amount of labeled data.
This architecture computes self attention in local windows and has shown better performance in comparison to ViT. In addition, the hierarchical nature of Swin Transformers makes them well suited for tasks requiring multiscale modeling.
Following the success of the pioneering UNETR model with a ViT-based encoder that directly uses 3D patch embeddings, Swin UNETR uses a 3D Swin Transformer encoder with a pyramid-like architecture.
In the encoder of the Swin UNETR, self-attention is computed in local windows since computing naive global self-attention is not feasible for high-resolution feature maps. In order to increase the receptive field beyond the local windows, window-shifting is used to compute the region interaction for different windows.
The encoder of the Swin UNETR is connected to a residual UNet-like decoder at five different resolutions by skip connections. It can capture multiscale feature representations for dense prediction tasks, such as medical image segmentation.
Swin UNETR model performance
After fine-tuning with the Beyond the Cranial Vault (BTCV) Segmentation Challenge on 13 abdominal organs in CT and the segmentation tasks from the Medical Segmentation Decathlon (MSD) dataset, the model achieved state-of-the-art accuracy on the public leaderboards.
BTCV
In the BTCV, Swin UNETR obtained an average Dice of 0.918, outperforming other top-ranked models.
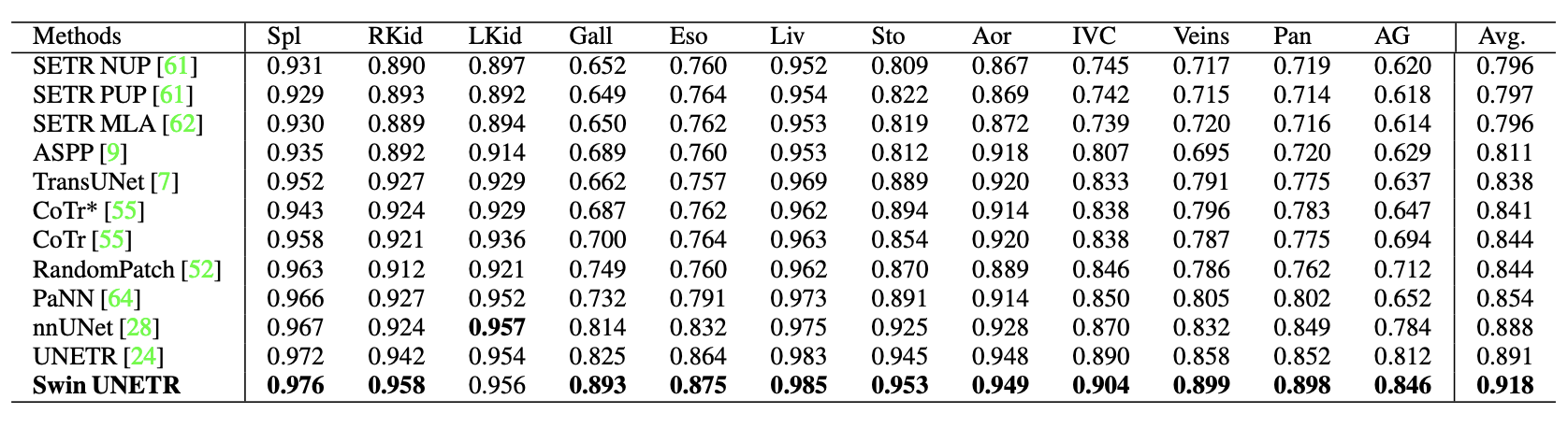
There are improvements compared to prior state-of-the-art methods for smaller organs, such as the splenic and portal veins (3.6%), pancreas (1.6%), and adrenal glands (3.8%.) Small organ data label segmentation is an excruciatingly difficult task for a radiologist. The improvement can be seen in the figure below.
MSD
In the MSD, Swin UNETR achieved state-of-the-art performance in brain tumor, lung, pancreas, and colon. The results are comparable for the heart, liver, hippocampus, prostate, hepatic vessel, and spleen. Overall, Swin UNETR presented the best average Dice of 78.68% across all 10 tasks and achieved the top ranking on the MSD leaderboard.
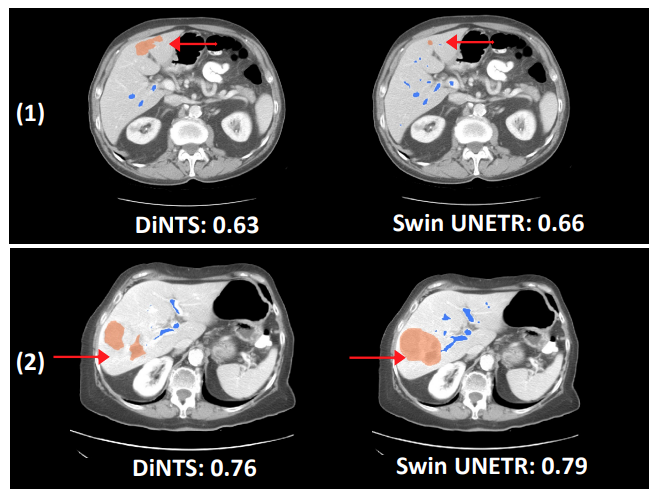
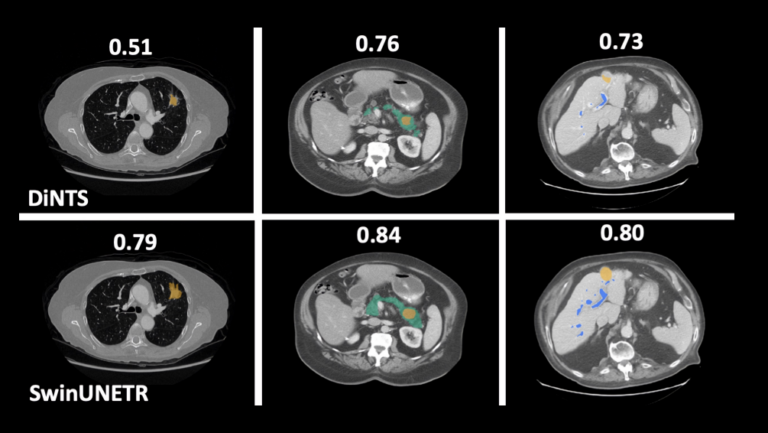
Swin UNETR has shown better segmentation performance using significantly fewer training GPU hours compared to DiNTS—a powerful AutoML methodology for medical image segmentation. For instance, qualitative segmentation outputs for the task of hepatic vessel segmentation demonstrate the capability of Swin UNETR to better model the long-range spatial dependencies.
Conclusion
The Swin UNETR architecture provides a much-needed breakthrough in medical imaging using transformers. Given the need in medical imaging to build accurate models quickly, with Swin UNETR data scientists can pretrain on a large corpus of unlabeled data. This reduces cost and time associated with expert annotation by radiologists, pathologists, and other clinical teams. Here we show SOTA segmentation performance which are used for organ detection and automatic volume measurements.
To learn more:
- Check out this work at the CVPR conference.
- Read the study Self-Supervised Pre-Training of Swin Transformers for 3D Medical Image Analysis.
- Download the SwinUNETR code on GitHub.
