 Developers have shown a lot of excitement for NVIDIA NIM microservices, a set of easy-to-use cloud-native microservices that shortens the time-to-market and…
Developers have shown a lot of excitement for NVIDIA NIM microservices, a set of easy-to-use cloud-native microservices that shortens the time-to-market and…
Developers have shown a lot of excitement for NVIDIA NIM microservices, a set of easy-to-use cloud-native microservices that shortens the time-to-market and simplifies the deployment of generative AI models anywhere, across cloud, data centers, cloud, and GPU-accelerated workstations. To meet the demands of diverse use cases, NVIDIA is bringing to market a variety of different AI models…

 Llama 3.1 Nemotron 70B Reward model helps generate high-quality training data that aligns with human preferences for finance, retail, healthcare, scientific…
Llama 3.1 Nemotron 70B Reward model helps generate high-quality training data that aligns with human preferences for finance, retail, healthcare, scientific…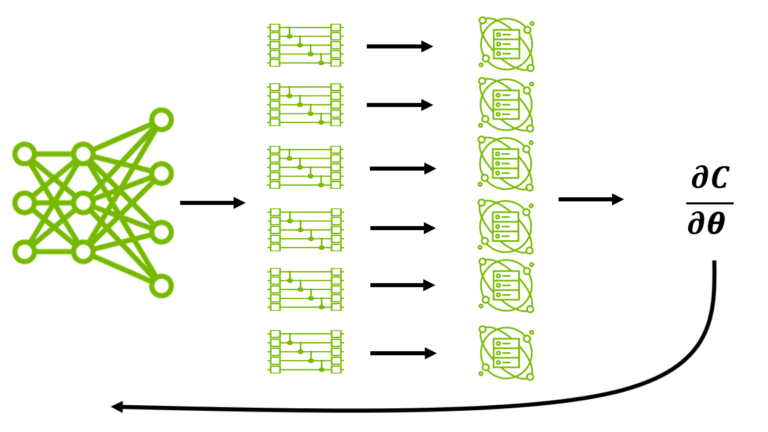 AI techniques like large language models (LLMs) are rapidly transforming many scientific disciplines. Quantum computing is no exception. A collaboration between…
AI techniques like large language models (LLMs) are rapidly transforming many scientific disciplines. Quantum computing is no exception. A collaboration between… Some of Africa’s most resource-constrained farmers are gaining access to on-demand, AI-powered advice through a multimodal chatbot that gives detailed…
Some of Africa’s most resource-constrained farmers are gaining access to on-demand, AI-powered advice through a multimodal chatbot that gives detailed… The new release includes several new features including improved stdpar programming and Arm processor support.
The new release includes several new features including improved stdpar programming and Arm processor support. Many of the most exciting applications of large language models (LLMs), such as interactive speech bots, coding co-pilots, and search, need to begin responding…
Many of the most exciting applications of large language models (LLMs), such as interactive speech bots, coding co-pilots, and search, need to begin responding…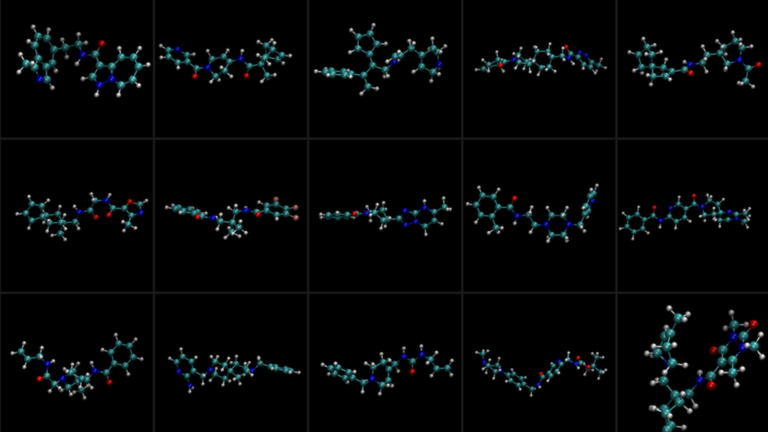 Drug discovery aims to develop new therapeutic agents that effectively target diseases while minimizing side effects for patients. Using multimodal data—such…
Drug discovery aims to develop new therapeutic agents that effectively target diseases while minimizing side effects for patients. Using multimodal data—such…