Every week, GFN Thursday brings new games to the cloud, featuring some of the latest and greatest titles for members to play. Leading the seven games joining GeForce NOW this week is the newest game in Ninja Theory’s Hellblade franchise, Senua’s Saga: Hellblade II. This day-and-date release expands the cloud gaming platform’s extensive library of
Read Article
Leading German automotive technology company enhances manufacturing workflows with OpenUSD-powered virtual factory solutions.
NVIDIA today reported revenue for the first quarter ended April 28, 2024, of $26.0 billion, up 18% from the previous quarter and up 262% from a year ago.
Just Released: NVIDIA HPC SDK 24.5
 NVIDIA HPC SDK 24.5 updates include support for new NVPL components and CUDA 12.4.
NVIDIA HPC SDK 24.5 updates include support for new NVPL components and CUDA 12.4.
NVIDIA HPC SDK 24.5 updates include support for new NVPL components and CUDA 12.4.
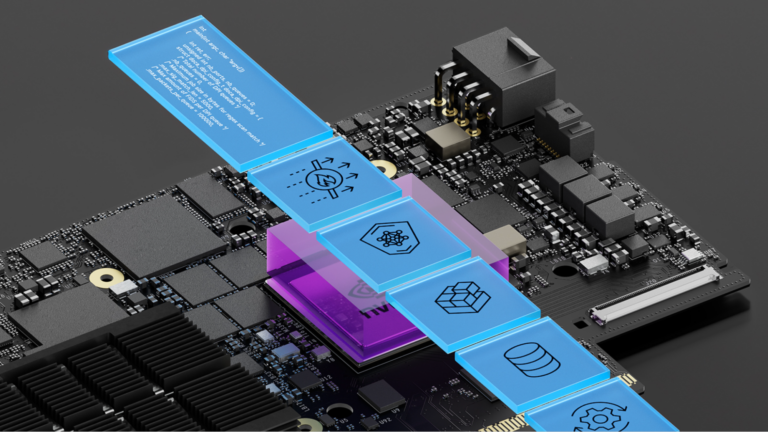 The NVIDIA DOCA acceleration framework empowers developers with extensive libraries, drivers, and APIs to create high-performance applications and services for…
The NVIDIA DOCA acceleration framework empowers developers with extensive libraries, drivers, and APIs to create high-performance applications and services for…
The NVIDIA DOCA acceleration framework empowers developers with extensive libraries, drivers, and APIs to create high-performance applications and services for NVIDIA BlueField DPUs and SuperNICs. DOCA 2.7 is a comprehensive, feature-rich release that further underpins the scope and value of the DOCA software framework. It offers several new libraries, turn-key applications…
In the vector space of all advice…
Just Released: Nsight Compute 2024.2
 Nsight Compute 2024.2 adds Python syntax highlighting and call stacks, a redesigned report header, and source page statistics to make CUDA optimization easier.
Nsight Compute 2024.2 adds Python syntax highlighting and call stacks, a redesigned report header, and source page statistics to make CUDA optimization easier.
Nsight Compute 2024.2 adds Python syntax highlighting and call stacks, a redesigned report header, and source page statistics to make CUDA optimization easier.
Just Released: CUDA Toolkit 12.5
 CUDA Toolkit 12.5 supports new NVIDIA L20 and H20 GPUs and simultaneous compute and graphics to DirectX, and updates Nsight Compute and CUDA-X Libraries.
CUDA Toolkit 12.5 supports new NVIDIA L20 and H20 GPUs and simultaneous compute and graphics to DirectX, and updates Nsight Compute and CUDA-X Libraries.
CUDA Toolkit 12.5 supports new NVIDIA L20 and H20 GPUs and simultaneous compute and graphics to DirectX, and updates Nsight Compute and CUDA-X Libraries.
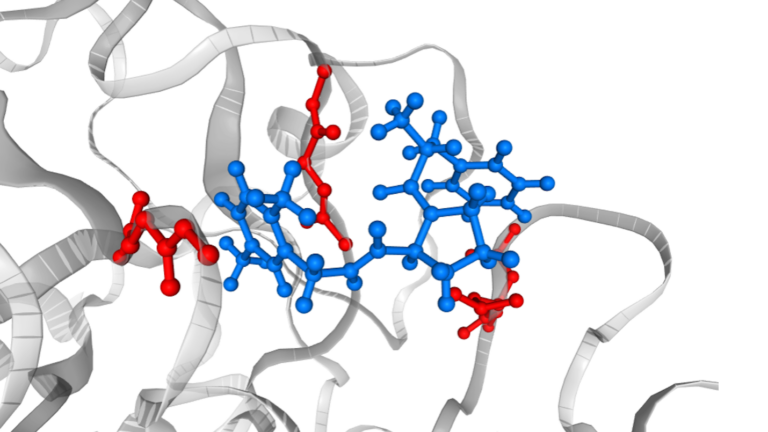 In drug discovery, approaches based on the so-called classical force field have been routinely used and considered useful. However, it is also widely recognized…
In drug discovery, approaches based on the so-called classical force field have been routinely used and considered useful. However, it is also widely recognized…
In drug discovery, approaches based on the so-called classical force field have been routinely used and considered useful. However, it is also widely recognized that some important physics are missing in the force field models, resulting in limited applicability. For example, force field models do not provide accurate predictions when comparing two molecules with different formal charges…
 Data curation is the first, and arguably the most important, step in the pretraining and continuous training of large language models (LLMs) and small language…
Data curation is the first, and arguably the most important, step in the pretraining and continuous training of large language models (LLMs) and small language…
Data curation is the first, and arguably the most important, step in the pretraining and continuous training of large language models (LLMs) and small language models (SLMs). NVIDIA recently announced the open-source release of NVIDIA NeMo Curator, a data curation framework that prepares large-scale, high-quality datasets for pretraining generative AI models. NeMo Curator, which is part of…
