 NVIDIA Modulus is now part of the NVIDIA AI Enterprise suite, supporting PyTorch 2.0, CUDA 12, and new samples.
NVIDIA Modulus is now part of the NVIDIA AI Enterprise suite, supporting PyTorch 2.0, CUDA 12, and new samples.
NVIDIA Modulus is now part of the NVIDIA AI Enterprise suite, supporting PyTorch 2.0, CUDA 12, and new samples.
 NVIDIA Modulus is now part of the NVIDIA AI Enterprise suite, supporting PyTorch 2.0, CUDA 12, and new samples.
NVIDIA Modulus is now part of the NVIDIA AI Enterprise suite, supporting PyTorch 2.0, CUDA 12, and new samples.
NVIDIA Modulus is now part of the NVIDIA AI Enterprise suite, supporting PyTorch 2.0, CUDA 12, and new samples.

The last few years have seen rapid progress in systems that can automatically process complex business documents and turn them into structured objects. A system that can automatically extract data from documents, e.g., receipts, insurance quotes, and financial statements, has the potential to dramatically improve the efficiency of business workflows by avoiding error-prone, manual work. Recent models, based on the Transformer architecture, have shown impressive gains in accuracy. Larger models, such as PaLM 2, are also being leveraged to further streamline these business workflows. However, the datasets used in academic literature fail to capture the challenges seen in real-world use cases. Consequently, academic benchmarks report strong model accuracy, but these same models do poorly when used for complex real-world applications.
In “VRDU: A Benchmark for Visually-rich Document Understanding”, presented at KDD 2023, we announce the release of the new Visually Rich Document Understanding (VRDU) dataset that aims to bridge this gap and help researchers better track progress on document understanding tasks. We list five requirements for a good document understanding benchmark, based on the kinds of real-world documents for which document understanding models are frequently used. Then, we describe how most datasets currently used by the research community fail to meet one or more of these requirements, while VRDU meets all of them. We are excited to announce the public release of the VRDU dataset and evaluation code under a Creative Commons license.
First, we compared state-of-the-art model accuracy (e.g., with FormNet and LayoutLMv2) on real-world use cases to academic benchmarks (e.g., FUNSD, CORD, SROIE). We observed that state-of-the-art models did not match academic benchmark results and delivered much lower accuracy in the real world. Next, we compared typical datasets for which document understanding models are frequently used with academic benchmarks and identified five dataset requirements that allow a dataset to better capture the complexity of real-world applications:
 |
The VRDU dataset is a combination of two publicly available datasets, Registration Forms and Ad-Buy forms. These datasets provide examples that are representative of real-world use cases, and satisfy the five benchmark requirements described above.
The Ad-buy Forms dataset consists of 641 documents with political advertisement details. Each document is either an invoice or receipt signed by a TV station and a campaign group. The documents use tables, multi-columns, and key-value pairs to record the advertisement information, such as the product name, broadcast dates, total price, and release date and time.
The Registration Forms dataset consists of 1,915 documents with information about foreign agents registering with the US government. Each document records essential information about foreign agents involved in activities that require public disclosure. Contents include the name of the registrant, the address of related bureaus, the purpose of activities, and other details.
We gathered a random sample of documents from the public Federal Communications Commission (FCC) and Foreign Agents Registration Act (FARA) sites, and converted the images to text using Google Cloud’s OCR. We discarded a small number of documents that were several pages long and the processing did not complete in under two minutes. This also allowed us to avoid sending very long documents for manual annotation — a task that can take over an hour for a single document. Then, we defined the schema and corresponding labeling instructions for a team of annotators experienced with document-labeling tasks.
The annotators were also provided with a few sample labeled documents that we labeled ourselves. The task required annotators to examine each document, draw a bounding box around every occurrence of an entity from the schema for each document, and associate that bounding box with the target entity. After the first round of labeling, a pool of experts were assigned to review the results. The corrected results are included in the published VRDU dataset. Please see the paper for more details on the labeling protocol and the schema for each dataset.
 |
| Existing academic benchmarks (FUNSD, CORD, SROIE, Kleister-NDA, Kleister-Charity, DeepForm) fall-short on one or more of the five requirements we identified for a good document understanding benchmark. VRDU satisfies all of them. See our paper for background on each of these datasets and a discussion on how they fail to meet one or more of the requirements. |
We built four different model training sets with 10, 50, 100, and 200 samples respectively. Then, we evaluated the VRDU datasets using three tasks (described below): (1) Single Template Learning, (2) Mixed Template Learning, and (3) Unseen Template Learning. For each of these tasks, we included 300 documents in the testing set. We evaluate models using the F1 score on the testing set.
The objective is to be able to evaluate models on their data efficiency. In our paper, we compared two recent models using the STL, MTL, and UTL tasks and made three observations. First, unlike with other benchmarks, VRDU is challenging and shows that models have plenty of room for improvements. Second, we show that few-shot performance for even state-of-the-art models is surprisingly low with even the best models resulting in less than an F1 score of 0.60. Third, we show that models struggle to deal with structured repeated fields and perform particularly poorly on them.
We release the new Visually Rich Document Understanding (VRDU) dataset that helps researchers better track progress on document understanding tasks. We describe why VRDU better reflects practical challenges in this domain. We also present experiments showing that VRDU tasks are challenging, and recent models have substantial headroom for improvements compared to the datasets typically used in the literature with F1 scores of 0.90+ being typical. We hope the release of the VRDU dataset and evaluation code helps research teams advance the state of the art in document understanding.
Many thanks to Zilong Wang, Yichao Zhou, Wei Wei, and Chen-Yu Lee, who co-authored the paper along with Sandeep Tata. Thanks to Marc Najork, Riham Mansour and numerous partners across Google Research and the Cloud AI team for providing valuable insights. Thanks to John Guilyard for creating the animations in this post.
 NVIDIA Nsight Developer Tools provide comprehensive access to NVIDIA GPUs and graphics APIs for performance analysis, optimization, and debugging activities….
NVIDIA Nsight Developer Tools provide comprehensive access to NVIDIA GPUs and graphics APIs for performance analysis, optimization, and debugging activities….
NVIDIA Nsight Developer Tools provide comprehensive access to NVIDIA GPUs and graphics APIs for performance analysis, optimization, and debugging activities. When using advanced rendering techniques like ray tracing or path tracing, Nsight tools are your companion for creating a smooth and polished experience.
At SIGGRAPH 2023, NVIDIA hosted a lab exploring how to use NVIDIA Nsight Tools to debug and profile ray tracing applications. New versions of the NVIDIA Nsight Aftermath SDK, NVIDIA Nsight Graphics, and NVIDIA Nsight Systems are also available. For more information on Nsight Tools released at SIGGRAPH, check out the latest video on NVIDIA Graphics Tools.
This post explores how Nsight Aftermath SDK 2023.2 speeds up GPU crash debugging with improved event marker performance.
Few issues are as pressing as a GPU crash, which can abruptly block development progress until resolved. Developers and end users alike find these crashes frustrating, especially when they can’t capture useful debugging information from the GPU pipeline at the moment of failure. To shed light on hidden exceptions, the Nsight Aftermath SDK opens a window into the GPU at the moment a game fails. This helps pinpoint the source of the issue and guides the developer in resolving it.
The Nsight Aftermath SDK generates GPU crash dump files that load into NVIDIA Nsight Graphics to visualize the GPU state—revealing MMU fault information, warp details, problematic shader source, and more. Integrating Aftermath into existing crash reporters also provides more granular pipeline dumps from end-users’ machines, providing actionable reports. Today’s update to the Nsight Aftermath SDK improves the contextual data provided through low-overhead, application-specific markers.
Event marker performance has been enhanced in the Nsight Aftermath SDK for DirectX 12 applications. You can insert these markers into your CPU code at desired intervals, and the significantly reduced overhead makes them usable in shipping applications. Markers are written to the Aftermath crash dump file, indicating where in the application’s frame a GPU exception occurred. With this information, you can determine the workload executing on the GPU and view what shaders were in use at the time of the crash.
The 2023.2 version of the Nsight Aftermath SDK also supports collecting and displaying shader register values to aid in debugging streaming multiprocessor (SM) exceptions. On the SM, registers store the results of instructions as they are executing. This data is particularly relevant to determining the source of a crash if a shader workload triggered the failure. After being written to an Nsight Aftermath dump file, you can inspect the register values for faulting threads in Nsight Graphics. This helps you determine where and why the shader execution failed.
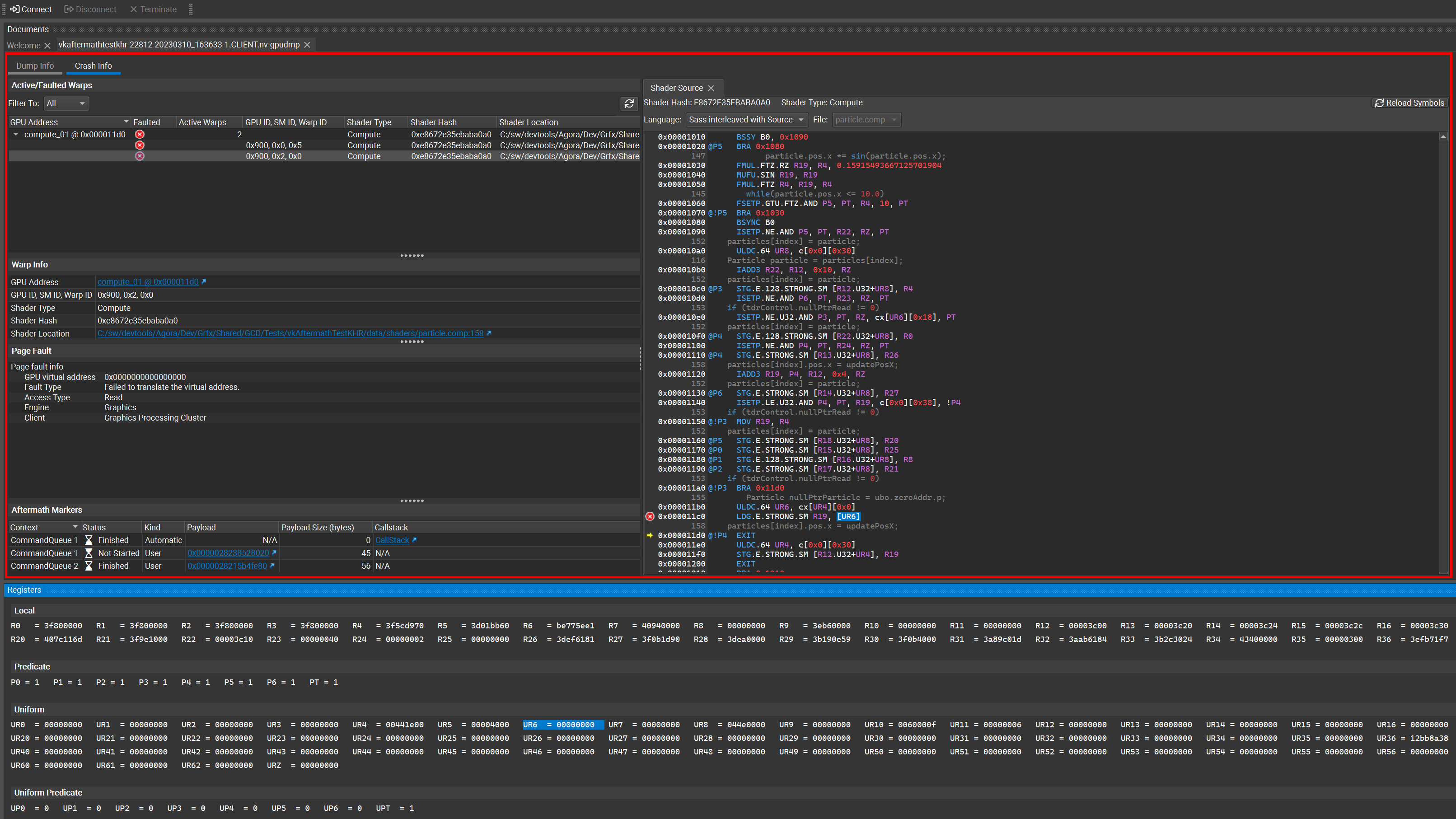
SM register data is now available for DirectX 12 and Vulkan applications. Note that viewing this data requires NVIDIA Nsight Graphics Pro. Coordinate with your NVIDIA Developer Technologies or Developer Relations contact, or reach out, to request access.
Nsight Aftermath is also now compatible with the latest applications using cutting-edge DirectX12 features through the DirectX Agility SDK.
Getting started with the SDK is easy. Here are some tips to help you use GPU crash dumps and event markers. More information is included in the Read Me section of the download.
GFSDK_Aftermath_EnableGpuCrashDumps. Note that crash dumps won’t be made for devices generated before that call. Make sure it’s enabled first.Tip: To use event markers, make sure that the event marker flag is enabled at this step. You can also use the Nsight Aftermath Monitor application to enable SM register collection.
Tip: the Aftermath API provides a simple and lightweight solution for inserting event markers on the GPU timeline. To keep CPU overhead to a minimum, you can set dataSize=0 to instruct Aftermath to rely on the application to manage and resolve marker data itself.
Download all of the new Nsight Developer Tools announced at SIGGRAPH.
Dive deeper or ask questions in Developer Tools forums or learn more about graphics development with Nsight Tools at SIGGRAPH 2023.
 NVIDIA is providing developers with an advanced platform to create scalable, branded, custom extended reality (XR) products with the new NVIDIA CloudXR Suite….
NVIDIA is providing developers with an advanced platform to create scalable, branded, custom extended reality (XR) products with the new NVIDIA CloudXR Suite….
NVIDIA is providing developers with an advanced platform to create scalable, branded, custom extended reality (XR) products with the new NVIDIA CloudXR Suite.
Built on a new architecture, NVIDIA CloudXR Suite is a major step forward in scaling the XR ecosystem. It provides a platform for developers, professionals, and enterprise teams to flexibly orchestrate and scale XR workloads across operating systems, including virtual machines in Windows and Linux-based systems such as containers.
With the NVIDIA CloudXR streaming stack, users can build flexible, high-performance cloud solutions capable of streaming the most demanding immersive experiences. Teams can also access NVIDIA streaming technology to effectively manage the quality of the streaming across large public and private networks, including the internet.
Immersive content developers have a challenge in supporting both tethered devices, driven by high-powered graphics cards, and mobile devices that have limited graphics power.
Using NVIDIA CloudXR, developers can create high-quality versions of applications—built to take advantage of powerful GPUs—and still target users with mobile XR devices using NVIDIA CloudXR streaming.
Additionally, cloud service providers (CSPs), orchestrators, and system integrators can extend GPU services with interactive graphics to support next-generation XR applications.
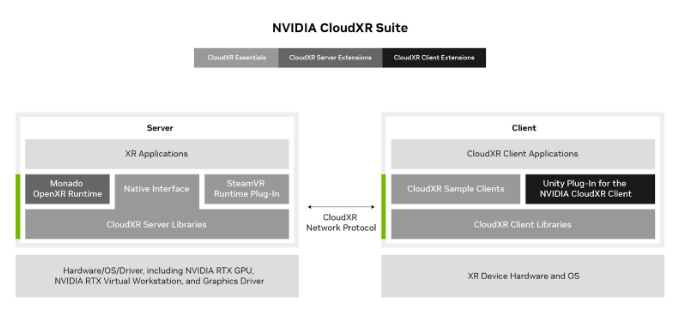
NVIDIA CloudXR Suite is composed of three components: CloudXR Essentials, CloudXR Server Extensions, and CloudXR Client Extensions.
CloudXR Essentials provide the underlying streaming layer, complete with new improvements such as 5G L4S optimizations, QoS algorithms, and enhanced logging tools. Essentials also includes the SteamVR plug-in, along with sample clients and a new server-side API that can be directly integrated into XR applications. This removes the need for a separate XR runtime.
CloudXR Server Extensions extend server-side interfaces with a source code addition to Collabara’s Monado OpenXR runtime. The new CloudXR Server API contained in CloudXR Essentials, plus the OpenXR API, represent the gateway to scaling XR distribution for orchestration partners.
CloudXR Client Extensions empower users to build custom CloudXR client applications. The first offering is a Unity plug-in for CloudXR. With this plug-in, Unity app developers can more easily build applications with branded custom interfaces and lobbies before connecting to the CloudXR streaming server.
“The new CloudXR Server Extensions bring more opportunities for software developers to use Monado’s OpenXR runtime to build next generation immersive experiences,” said Frédéric Plourde, XR Lead at Collabora.
PureWeb helps organizations across industries adopt real-time 3D technology to improve operations. Using NVIDIA CloudXR, PureWeb shows customers how to deliver complex, immersive XR workloads at scale, on their current devices.
“We want to provide our customers with access to the GPU resources and streaming technologies they need, so they can share immersive experiences without worrying about not having enough computing resources,” said Chris Jarabek, VP of Product Development at PureWeb. “With this new advancement through CloudXR Suite, we can better scale with OpenXR and build with that.”
The team at Innoactive has integrated NVIDIA CloudXR into their VR application deployment platform, Innoactive Portal. Many Innoactive customers are using the platform to provide high-quality immersive training to users, wherever they are.
“Many of our customers are building applications and plan to stream them to all-in-one headsets or other mobile XR devices,” said Daniel Seidl, CEO of Innoactive. “With the Unity plug-in, our customers can now stream from AWS and Microsoft Azure with CloudXR, making XR streaming even more accessible from the cloud.”

Adaptive computation refers to the ability of a machine learning system to adjust its behavior in response to changes in the environment. While conventional neural networks have a fixed function and computation capacity, i.e., they spend the same number of FLOPs for processing different inputs, a model with adaptive and dynamic computation modulates the computational budget it dedicates to processing each input, depending on the complexity of the input.
Adaptive computation in neural networks is appealing for two key reasons. First, the mechanism that introduces adaptivity provides an inductive bias that can play a key role in solving some challenging tasks. For instance, enabling different numbers of computational steps for different inputs can be crucial in solving arithmetic problems that require modeling hierarchies of different depths. Second, it gives practitioners the ability to tune the cost of inference through greater flexibility offered by dynamic computation, as these models can be adjusted to spend more FLOPs processing a new input.
Neural networks can be made adaptive by using different functions or computation budgets for various inputs. A deep neural network can be thought of as a function that outputs a result based on both the input and its parameters. To implement adaptive function types, a subset of parameters are selectively activated based on the input, a process referred to as conditional computation. Adaptivity based on the function type has been explored in studies on mixture-of-experts, where the sparsely activated parameters for each input sample are determined through routing.
Another area of research in adaptive computation involves dynamic computation budgets. Unlike in standard neural networks, such as T5, GPT-3, PaLM, and ViT, whose computation budget is fixed for different samples, recent research has demonstrated that adaptive computation budgets can improve performance on tasks where transformers fall short. Many of these works achieve adaptivity by using dynamic depth to allocate the computation budget. For example, the Adaptive Computation Time (ACT) algorithm was proposed to provide an adaptive computational budget for recurrent neural networks. The Universal Transformer extends the ACT algorithm to transformers by making the computation budget dependent on the number of transformer layers used for each input example or token. Recent studies, like PonderNet, follow a similar approach while improving the dynamic halting mechanisms.
In the paper “Adaptive Computation with Elastic Input Sequence”, we introduce a new model that utilizes adaptive computation, called AdaTape. This model is a Transformer-based architecture that uses a dynamic set of tokens to create elastic input sequences, providing a unique perspective on adaptivity in comparison to previous works. AdaTape uses an adaptive tape reading mechanism to determine a varying number of tape tokens that are added to each input based on input’s complexity. AdaTape is very simple to implement, provides an effective knob to increase the accuracy when needed, but is also much more efficient compared to other adaptive baselines because it directly injects adaptivity into the input sequence instead of the model depth. Finally, Adatape offers better performance on standard tasks, like image classification, as well as algorithmic tasks, while maintaining a favorable quality and cost tradeoff.
AdaTape uses both the adaptive function types and a dynamic computation budget. Specifically, for a batch of input sequences after tokenization (e.g., a linear projection of non-overlapping patches from an image in the vision transformer), AdaTape uses a vector representing each input to dynamically select a variable-sized sequence of tape tokens.
AdaTape uses a bank of tokens, called a “tape bank”, to store all the candidate tape tokens that interact with the model through the adaptive tape reading mechanism. We explore two different methods for creating the tape bank: an input-driven bank and a learnable bank.
The general idea of the input-driven bank is to extract a bank of tokens from the input while employing a different approach than the original model tokenizer for mapping the raw input to a sequence of input tokens. This enables dynamic, on-demand access to information from the input that is obtained using a different point of view, e.g., a different image resolution or a different level of abstraction.
In some cases, tokenization in a different level of abstraction is not possible, thus an input-driven tape bank is not feasible, such as when it’s difficult to further split each node in a graph transformer. To address this issue, AdaTape offers a more general approach for generating the tape bank by using a set of trainable vectors as tape tokens. This approach is referred to as the learnable bank and can be viewed as an embedding layer where the model can dynamically retrieve tokens based on the complexity of the input example. The learnable bank enables AdaTape to generate a more flexible tape bank, providing it with the ability to dynamically adjust its computation budget based on the complexity of each input example, e.g., more complex examples retrieve more tokens from the bank, which let the model not only use the knowledge stored in the bank, but also spend more FLOPs processing it, since the input is now larger.
Finally, the selected tape tokens are appended to the original input and fed to the following transformer layers. For each transformer layer, the same multi-head attention is used across all input and tape tokens. However, two different feed-forward networks (FFN) are used: one for all tokens from the original input and the other for all tape tokens. We observed slightly better quality by using separate feed-forward networks for input and tape tokens.
We evaluate AdaTape on parity, a very challenging task for the standard Transformer, to study the effect of inductive biases in AdaTape. With the parity task, given a sequence 1s, 0s, and -1s, the model has to predict the evenness or oddness of the number of 1s in the sequence. Parity is the simplest non-counter-free or periodic regular language, but perhaps surprisingly, the task is unsolvable by the standard Transformer.
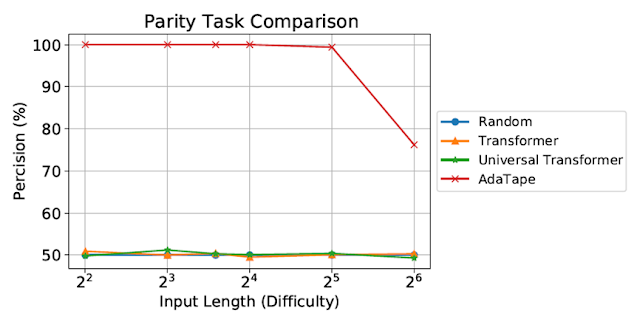 |
| Evaluation on the parity task. The standard Transformer and Universal Transformer were unable to perform this task, both showing performance at the level of a random guessing baseline. |
Despite being evaluated on short, simple sequences, both the standard Transformer and Universal Transformers were unable to perform the parity task as they are unable to maintain a counter within the model. However, AdaTape outperforms all baselines, as it incorporates a lightweight recurrence within its input selection mechanism, providing an inductive bias that enables the implicit maintenance of a counter, which is not possible in standard Transformers.
We also evaluate AdaTape on the image classification task. To do so, we trained AdaTape on ImageNet-1K from scratch. The figure below shows the accuracy of AdaTape and the baseline methods, including A-ViT, and the Universal Transformer ViT (UViT and U2T) versus their speed (measured as number of images, processed by each code, per second). In terms of quality and cost tradeoff, AdaTape performs much better than the alternative adaptive transformer baselines. In terms of efficiency, larger AdaTape models (in terms of parameter count) are faster than smaller baselines. Such results are consistent with the finding from previous work that shows that the adaptive model depth architectures are not well suited for many accelerators, like the TPU.
 |
| We evaluate AdaTape by training on ImageNet from scratch. For A-ViT, we not only report their results from the paper but also re-implement A-ViT by training from scratch, i.e., A-ViT(Ours). |
In addition to its performance on the parity task and ImageNet-1K, we also evaluated the token selection behavior of AdaTape with an input-driven bank on the JFT-300M validation set. To better understand the model’s behavior, we visualized the token selection results on the input-driven bank as heatmaps, where lighter colors mean that position is more frequently selected. The heatmaps reveal that AdaTape more frequently picks the central patches. This aligns with our prior knowledge, as central patches are typically more informative — especially in the context of datasets with natural images, where the main object is in the middle of the image. This result highlights the intelligence of AdaTape, as it can effectively identify and prioritize more informative patches to improve its performance.
 |
| We visualize the tape token selection heatmap of AdaTape-B/32 (left) and AdaTape-B/16 (right). The hotter / lighter color means the patch at this position is more frequently selected. |
AdaTape is characterized by elastic sequence lengths generated by the adaptive tape reading mechanism. This also introduces a new inductive bias that enables AdaTape to have the potential to solve tasks that are challenging for both standard transformers and existing adaptive transformers. By conducting comprehensive experiments on image recognition benchmarks, we demonstrate that AdaTape outperforms standard transformers and adaptive architecture transformers when computation is held constant.
One of the authors of this post, Mostafa Dehghani, is now at Google DeepMind.
Picture this: Creators can quickly create and customize 3D scene backgrounds with the help of generative AI, thanks to cutting-edge tools from Shutterstock. The visual-content provider is building services using NVIDIA Picasso — a cloud-based foundry for developing generative AI models for visual design. The work incorporates Picasso’s latest feature — announced today during NVIDIA Read article >
AI and accelerated computing were in the spotlight at SIGGRAPH — the world’s largest gathering of computer graphics experts — as NVIDIA founder and CEO Jensen Huang announced during his keynote address updates to NVIDIA Omniverse, a platform for building and connecting 3D tools and applications, as well as acceleration for Universal Scene Description (known as OpenUSD), the open and extensible ecosystem for 3D worlds.
New Omniverse Cloud APIs Help Developers Adopt OpenUSD; Generative AI Model ChatUSD LLM Converses in USD; RunUSD Translates USD to Interactive Graphics, DeepSearch LLM Enables Semantic 3D …
Professionals, teams, creators and others can tap into the power of AI to create high-quality audio and video effects — even using standard microphones and webcams — with the help of NVIDIA Maxine. The suite of GPU-accelerated software development kits and cloud-native microservices lets users deploy AI features that enhance audio, video and augmented-reality effects Read article >
Organizations across industries are using extended reality (XR) to redesign workflows and boost productivity, whether for immersive training or collaborative design reviews. With the growing use of all-in-one (AIO) headsets, more teams have adopted and integrated XR. While easing XR use, AIO headsets have modest compute and rendering power that can limit the graphics quality Read article >