This post defines NICs, SmartNICs, and lays out a cost-benefit analysis for NIC categories and use cases.
This post defines NICs, SmartNICs, and lays out a cost-benefit analysis for NIC categories and use cases.
This post was originally published on the Mellanox blog.
Everyone is talking about data processing unit–based SmartNICs but without answering one simple question: What is a SmartNIC and what do they do?
NIC stands for network interface card. Practically speaking, a NIC is a PCIe card that plugs into a server or storage box to enable connectivity to an Ethernet network. A DPU-based SmartNIC goes beyond simple connectivity and implements network traffic processing on the NIC that would necessarily be performed by the CPU, in the case of a foundational NIC.
Some vendors’ definitions of a DPU-based SmartNIC are focused entirely on the implementation. This is problematic, as different vendors have different architectures. Thus, a DPU-based SmartNIC can be ASIC–, FPGA–, and system-on-a-chip-based. Naturally, vendors who make just one kind of NIC insist that only their type of NIC should qualify as a SmartNIC.
- Excellent price performance
- Vendor development cost high
- Programmable and extensible
- Flexibility is limited to predefined capabilities

- Good performance but expensive
- Difficult to program
- Workload-specific optimization
- Good price performance
- C programmable processors
- Highest flexibility
- Easiest programmability
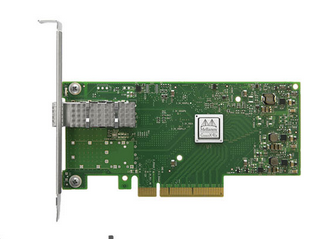
There are various tradeoffs between these different implementations with regards to cost, ease of programming, and flexibility. An ASIC is cost-effective and may deliver the best price performance, but it suffers from limited flexibility. An ASIC-based NIC, like the NVIDIA ConnectX-5, can have a programmable data path that is relatively simple to configure. Ultimately, that functionality has limits based on what functions are defined within the ASIC. That can prevent certain workloads from being supported.
By contrast, an FPGA NIC, such as the NVIDIA Innova-2 Flex, is highly programmable. With enough time and effort, it can be made to support almost any functionality relatively efficiently, within the constraints of the available gates. However, FPGAs are notoriously difficult to program and expensive.
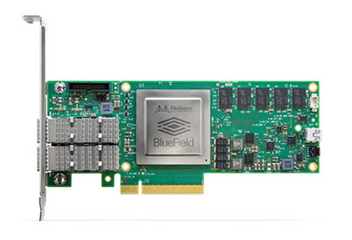
For more complex use cases, the SOC, such as the Mellanox BlueField DPU-programmable SmartNIC provides what appears to be the best DPU-based SmartNIC implementation option: good price performance, easy to program, and highly flexible.

Focusing on how a particular vendor implements a DPU-based SmartNIC doesn’t address what it’s capable of or how it should be architected. NVIDIA actually has products based on each of these architectures that could be classified as DPU-based SmartNICs. In fact, customers use each of these products for different workloads, depending on their needs. So the focus on implementation—ASIC vs. FPGA vs. SoC—reverses the ‘form follows function’ philosophy that underlies the best architectural achievements.
Rather than focusing on implementation, I tweaked this PC Magazine encyclopedia entry to give a working definition of what makes a NIC a DPU-based SmartNIC:
DPU-based SmartNIC:
A DPU-based network interface card (network adapter) that offloads processing tasks that the system CPU would normally handle. Using its own onboard processor, the DPU-based SmartNIC may be able to perform any combination of encryption/decryption, firewall, TCP/IP, and HTTP processing. SmartNICs are ideally suited for high-traffic web servers.
There are two things that I like about this definition. First, it focuses on the function more than the form. Second, it hints at this form with the statement, “…using its own onboard processor… to perform any combination of…” network processing tasks. So the embedded processor is key to achieving the flexibility to perform almost any networking function.
You could modernize that definition by adding that DPU-based SmartNICs might also perform network, storage, or GPU virtualization. Also, SmartNICs are also ideally suited for telco, security, machine learning, software-defined storage, and hyperconverged infrastructure servers, not just web servers.
NIC categories
Here’s how to differentiate three categories of NICs by the functions that the network adapters can support and use to accelerate different workloads:
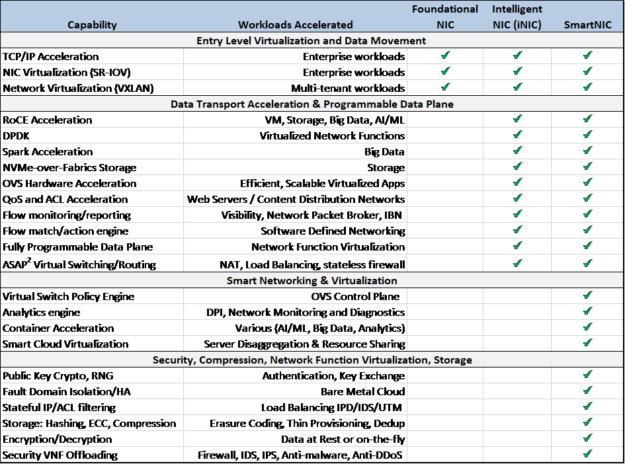
Here I’ve defined three categories of NICs, based on their ability to accelerate specific functionality:
- Foundational NIC
- Intelligent NIC (iNIC)
- DPU-based SmartNIC
The foundational, or basic NIC simply moves network traffic and has few or no offloads, other than possibly SRIOV and basic TCP acceleration. It doesn’t save any CPU cycles and can’t offload packet steering or traffic flows. At NVIDIA, we don’t even sell a foundational NIC any more.
The NVIDIA ConnectX adapter family features a programmable data path and accelerates a range of functions that first became important in public cloud use cases. For this reason, I’ve defined this type of NIC as an iNIC, although today on-premises enterprise, telco, and private clouds are just as likely as public cloud providers to need this type of programmability and acceleration functionality. Another name for it could be smarterNIC without the capital “S.”
In many cases, customers tell us they need DPU based SmartNIC capabilities that are being offered by a competitor with either an FPGA or a NIC combined with custom, proprietary processing engines. But when customers really look at the functions they need for their specific workloads, ultimately they decide that the ConnectX family of iNICs provides all the function, performance, and flexibility of other so-called SmartNICs at a fraction of the power and cost. So by the definition of SmartNIC that some competitors use – our ConnectX NICs are indeed SmartNICs, though we might call them intelligent NICs or smarter NICs. Our FPGA NIC (Innova) is also a SmartNIC in the classic sense, and our SoC NIC (using BlueField) is the smartest of SmartNICs, to the extent that we could call them Genius NICs
So, what is a SmartNIC? A DPU-based SmartNIC is a network adapter that accelerates functionality and offloads it from the server (or storage) CPU.
How you should build a DPU-based SmartNIC and which SmartNIC is the best for each workload… well, the devil is in the details. It’s important to dig into exactly what data path and virtualization accelerations are available and how they can be used. If you’re interested, see my next post, Achieving a Cloud-Scale Architecture with DPUs.
For more information about SmartNIC use cases, see the following resources:
- The Next Platform article: Living in the SmartNIC Future
- SDxCentral article: Smart NICs in White Boxes

 Learn how to simplify AI model deployment at the edge with NVIDIA Triton Inference Server on NVIDIA Jetson. Triton Inference Server is available on Jetson starting with the JetPack 4.6 release.
Learn how to simplify AI model deployment at the edge with NVIDIA Triton Inference Server on NVIDIA Jetson. Triton Inference Server is available on Jetson starting with the JetPack 4.6 release.
 The NVIDIA Deep Learning Institute offers free courses for all experience levels in deep learning, accelerated computing, and accelerated data science.
The NVIDIA Deep Learning Institute offers free courses for all experience levels in deep learning, accelerated computing, and accelerated data science. The early access version of the NVIDIA DOCA SDK was announced earlier this year at GTC. DOCA marks our focus on finding new ways to accelerate computing. The emergence of the DPU paradigm as the evolution of SmartNICs is finally here. We enable developers and application architects to squeeze more value out of general-purpose CPUs …
The early access version of the NVIDIA DOCA SDK was announced earlier this year at GTC. DOCA marks our focus on finding new ways to accelerate computing. The emergence of the DPU paradigm as the evolution of SmartNICs is finally here. We enable developers and application architects to squeeze more value out of general-purpose CPUs … 
 Recreate the past with the new NVIDIA Omniverse retro design challenge.
Recreate the past with the new NVIDIA Omniverse retro design challenge.